REGULATION OF MITOCHONDRIAL GENE EXPRESSION
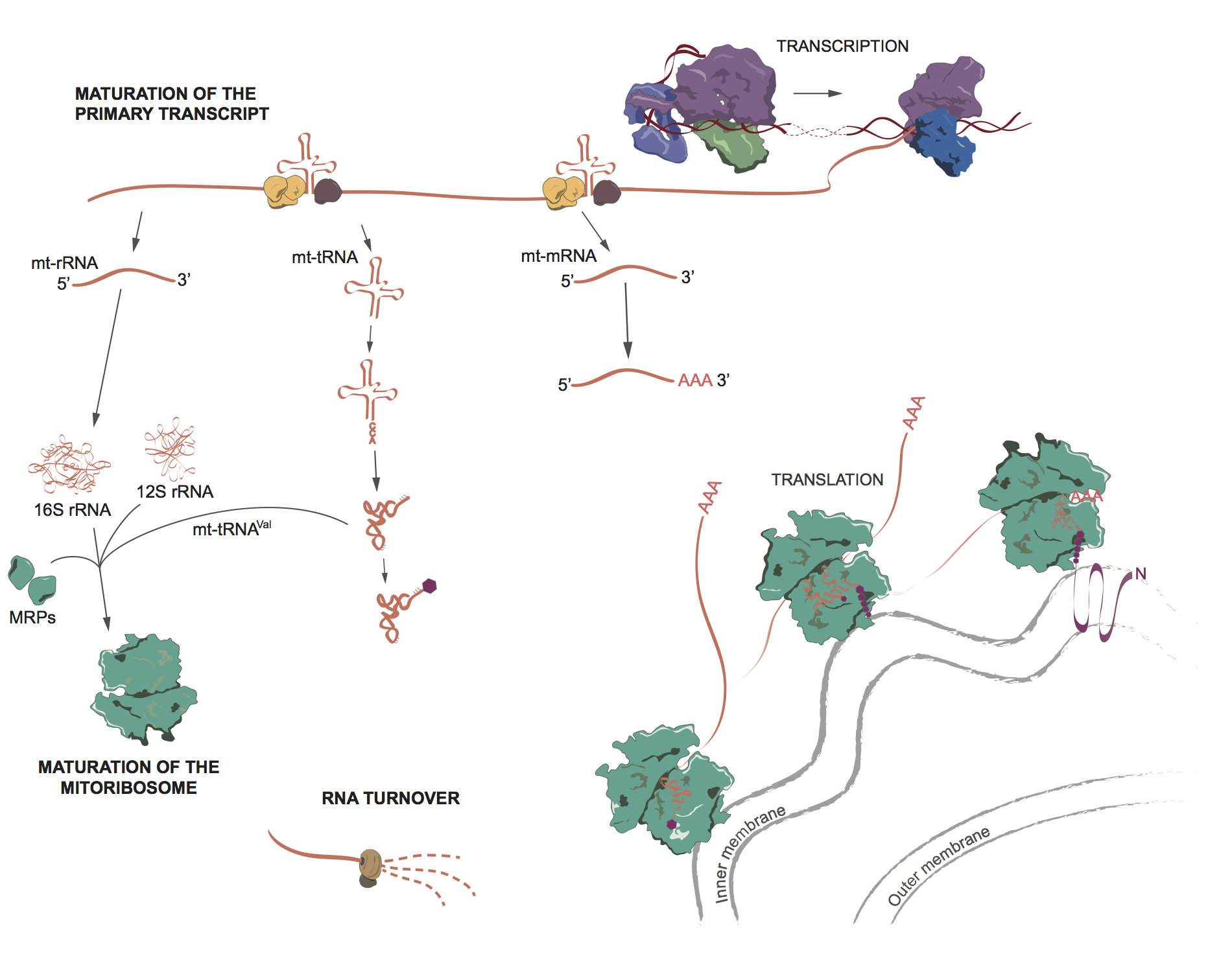
Figure | Mitochondrial RNA metabolism [1]
The human mitochondrial genome (mtDNA) encodes several structural components of the oxidative phosphorylation system (OxPhos) and RNA components for intramitochondrial protein synthesis. Therefore, mitochondria have evolved unique and highly specialised mechanisms to express genes contained within mtDNA [2].
The mitochondrial rRNAs (blue), mRNAs (red) and tRNAs (green) are transcribed as polycistronic units. Following the endonucleolytic processing, individual mRNA, rRNA and tRNA transcripts undergo post-transcriptional modifications. Several nucleotides of rRNAs are modified to facilitate mitoribosome biogenesis and function, a poly(A) tail is added to mRNAs and tRNA and rRNA undergo extensive post-transcriptional nucleotide modification, in addition to a CCA trinucleotide synthesis at the 3’ end, before being aminoacylated with a cognate amino acid. Decay and surveillance pathways have also been described for mammalian mtRNA [2] [3].
Establishing how defects in these processes contribute to human disease constitutes a major challenge. Recent years have brought many important insights into the regulation of mtDNA maintenance and expression. A number of novel factors have been identified, either by basic research approaches, or through the study of patients with respiratory chain disorders. The characterisation of these factors has furthered a basic understanding of their function in processes of mitochondrial DNA (mtDNA) replication, mtRNA metabolism and translation, and has also shed new light upon these processes. Nonetheless, many fundamental questions, frequently related to the pathology of mitochondrial diseases, remain unanswered and new questions have arisen.
Currently, we try to understand the regulation of mammalian mitochondrial gene expression. We focus mainly on studying fundamental processes of:
• Endonuclolytic processing of precursor mitochondrial RNA
• Polyadenylation of mitochondrial RNA [3] [4]
• Post-transcriptional modification (epitranscriptomics) of mitochondrial RNA [5] [6] [7] [8] [9]
• Mitoribosome biogenesis and function [10] [11] [12] [13]
REFERENCES
- D'Souza, AR, Minczuk, M (2018)
Mitochondrial transcription and translation: overview.
Essays Biochem 62, 309-320 - Rorbach, J, Minczuk, M (2012)
The post-transcriptional life of mammalian mitochondrial RNA.
Biochem J 444, 357-73 - Rorbach, J, Nicholls, TJJ, Minczuk, M (2011)
PDE12 removes mitochondrial RNA poly(A) tails and controls translation in human mitochondria.
Nucleic Acids Res 39, 7750-63 - Pearce, SF, Rorbach, J, Van Haute, L, D'Souza, AR, Rebelo-Guiomar, P, Powell, CA, Brierley, I, Firth, AE, Minczuk, M (2017)
Maturation of selected human mitochondrial tRNAs requires deadenylation.
eLife 6, 27596 - Rorbach, J, Boesch, P, Gammage, PA, Nicholls, TJJ, Pearce, SF, Patel, D, Hauser, A, Perocchi, F, Minczuk, M (2014)
MRM2 and MRM3 are involved in biogenesis of the large subunit of the mitochondrial ribosome.
Mol Biol Cell 25, 2542-55 - Van Haute, L, Dietmann, S, Kremer, L, Hussain, S, Pearce, SF, Powell, CA, Rorbach, J, Lantaff, R, Blanco, S, Sauer, S, Kotzaeridou, U, Hoffmann, GF, Memari, Y, Kolb-Kokocinski, A, Durbin, R, Mayr, JA, Frye, M, Prokisch, H, Minczuk, M (2016)
Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3.
Nat Commun 7, 12039 - Zaganelli, S, Rebelo-Guiomar, P, Maundrell, K, Rozanska, A, Pierredon, S, Powell, CA, Jourdain, AA, Hulo, N, Lightowlers, RN, Chrzanowska-Lightowlers, ZM, Minczuk, MA, Martinou, J-C (2017)
The pseudouridine synthase RPUSD4 is an essential component of mitochondrial RNA granules.
J Biol Chem 292, 4519-4532 - Powell, CA, Minczuk, M (2020)
TRMT2B is responsible for both tRNA and rRNA m 5 U-methylation in human mitochondria
RNA Biol 17, 451-462 - Rorbach, J, Gao, F, Powell, CA, D'Souza, A, Lightowlers, RN, Minczuk, MA, Chrzanowska-Lightowlers, ZM (2016)
Human mitochondrial ribosomes can switch their structural RNA composition.
Proc Natl Acad Sci U S A 113, 12198-12202 - Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V (2020)
Elongational stalling activates mitoribosome-associated quality control.
Science 370, 1105-1110 - D'Souza AR, Van Haute L, Powell CA, Mutti CD, Páleníková P, Rebelo-Guiomar P, Rorbach J, Minczuk M (2021)
YbeY is required for ribosome small subunit assembly and tRNA processing in human mitochondria.
- (2022)
A late-stage assembly checkpoint of the human mitochondrial ribosome large subunit.
Nat Commun. 13:929