MECHANISMS OF MITOPHAGY AND PINK1/PARKIN FUNCTION
PINK1 and Parkin (PRKN), two genes linked to Parkinson's disease, have been strongly linked to the selective degradation of mitochondria, termed mitophagy. The fact that loss of function mutations in these genes causes neurodegeneration establishes the importance of this process to neuronal survival. However, much of our current understanding comes from in vitro studies, so we still have a poor understanding of this process in a physiological context. For example, it is currently not clear to what extent turnover of mitochondrial components is via bulk autophagosome engulfment and degradation, versus small-scale vesicular trafficking. Nor is it clear what are the important molecular triggers for turnover; either the stimulus for initiating the PINK1/Parkin cascade or the critical Parkin targets.
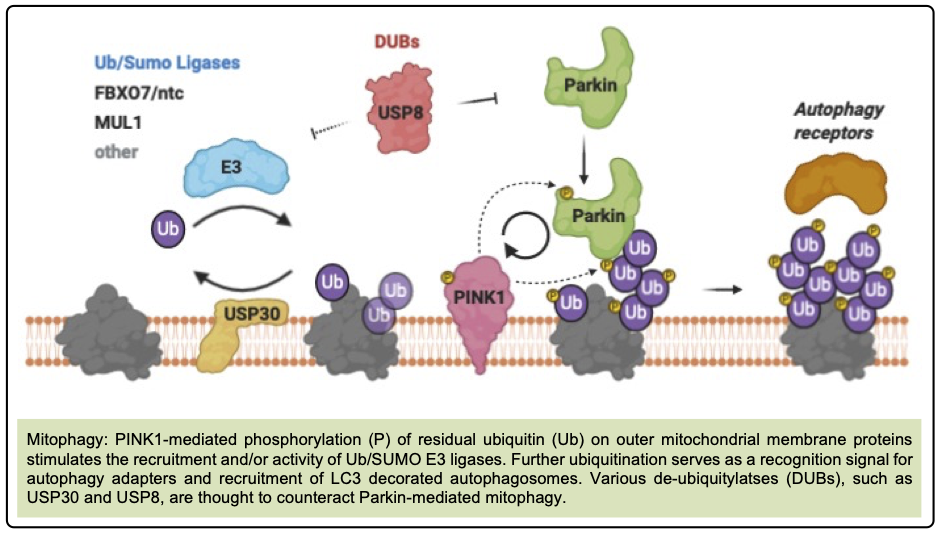
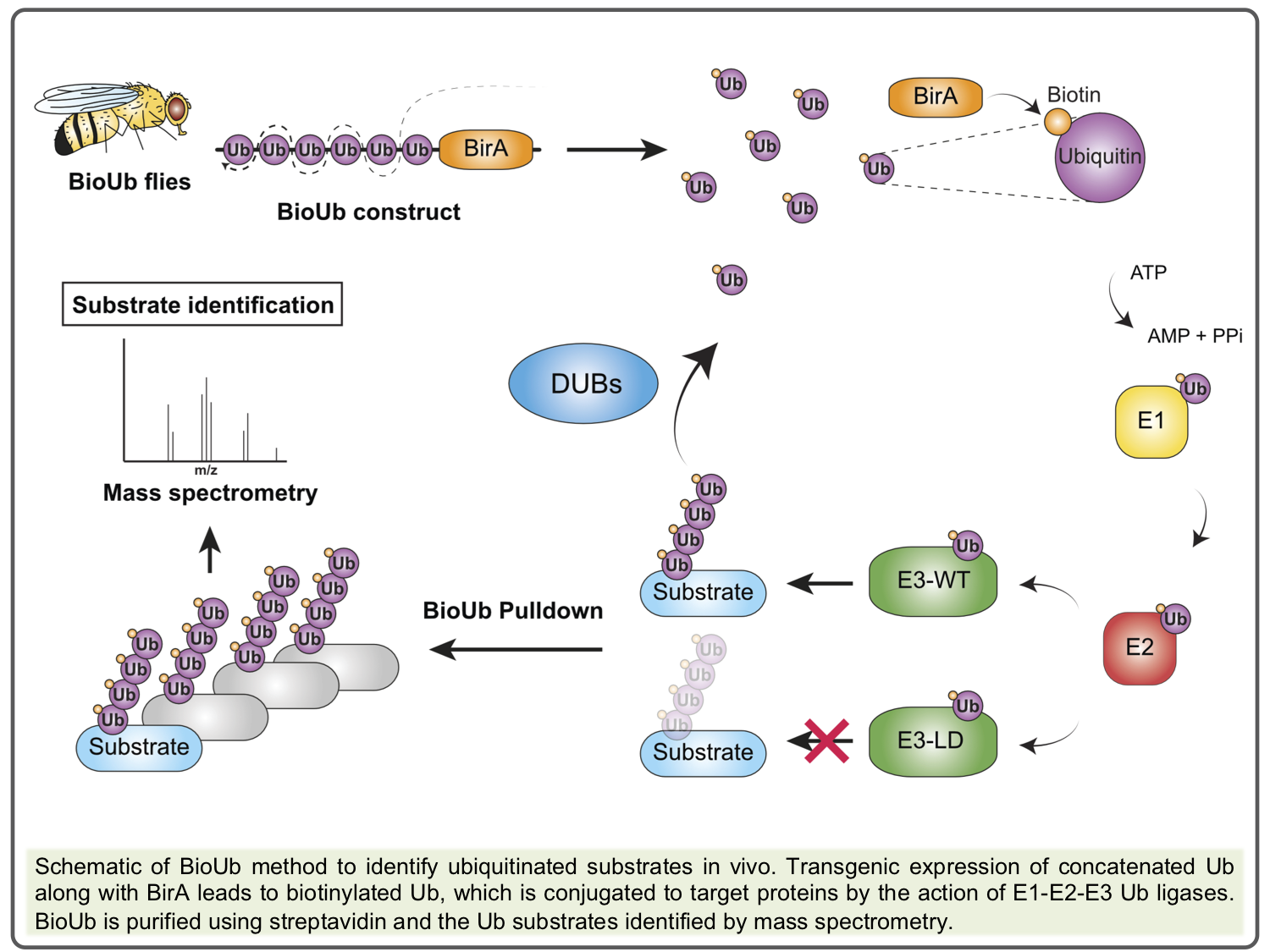
To address these questions we are using multiple molecular and genetic approaches to (i) monitor mitophagy events in vivo using recently published reporters [1], (ii) assess which mitochondrial stresses provoke mitophagy, (iii) identify the ubiquitination and SUMOylation targets of positive and negative regulators of mitophagy, e.g. Ub/SUMO ligases and deubiquitylases (DUBs)[2], especially USP30 [3],[4] and USP8[5], using the BioUb method[6],[7], (iv) determine which of these are critical regulators of the mitophagy process.
We are also analysing the potential role of immune signalling pathways, such as the STING pathway[8],[9], and defective immune activation in the pathological consequences of mutated PINK1/parkin.
REFERENCES
- Lee JJ, Sanchez-Martinez A, Zarate AMartinez, Benincá C, Mayor U, Clague MJ & Whitworth AJ (2018)
Basal mitophagy is widespread inbut minimally affected by loss of Pink1 or parkin.
J Cell Biol 217, 1613-1622 - Dikic I & Bremm A (2014)
DUBs counteract parkin for efficient mitophagy.
EMBO J 33, 2442-2443 - Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS & Sheng M (2014)
The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy.
Nature 510, 370-375 - Marcassa E, Kallinos A, Jardine J, Rusilowicz-Jones EV, Martinez A, Kuehl S, Islinger M, Clague MJ & Urbé S (2018)
Dual role of USP30 in controlling basal pexophagy and mitophagy.
EMBO Rep 19, e45595 - von Stockum S, Sanchez-Martinez A, Corrà S, Chakraborty J, Marchesan E, Locatello L, Da Re C, Cusumano P, Caicci F, Ferrari V, Costa R, Bubacco L, Rasotto MBerica, Szabó I, Whitworth AJ, Scorrano L & Ziviani E (2019)
Inhibition of the deubiquitinase USP8 corrects a Drosophila PINK1 model of mitochondria dysfunction.
Life Sci Alliance 2, e201900392 - Franco M, Seyfried NT, Brand AH, Peng J & Mayor U (2011)
A novel strategy to isolate ubiquitin conjugates reveals wide role for ubiquitination during neural development.
Mol Cell Proteomics 10, M110.002188 - Martinez A, Lectez B, Ramirez J, Popp O, Sutherland JD, Urbé S, Dittmar G, Clague MJ & Mayor U (2017)
Quantitative proteomic analysis of Parkin substrates in Drosophila neurons.
Mol Neurodegener 12, 29 - Lee JJ, Andreazza S & Whitworth AJ (2020)
The STING pathway does not contribute to behavioural or mitochondrial phenotypes in Drosophila Pink1/parkin or mtDNA mutator models.
Sci Rep 10, 2693 - Sliter DA, Martinez J, Hao L, Chen X, Sun N, Fischer TD, Burman JL, Li Y, Zhang Z, Narendra DP, Cai H, Borsche M, Klein C & Youle RJ (2018)
Parkin and PINK1 mitigate STING-induced inflammation.
Nature 561, 258-262