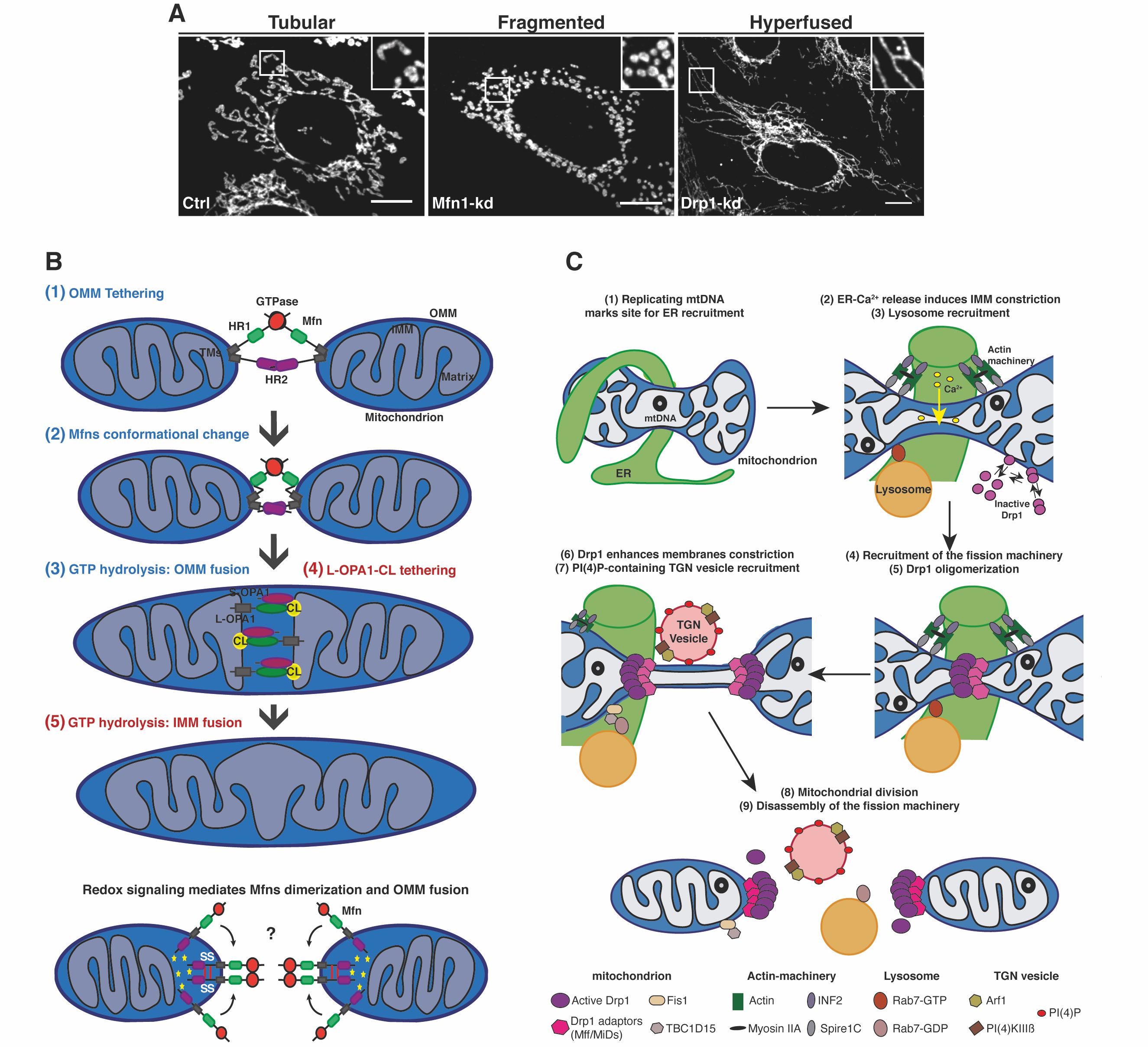
Figure 1. (A) Representative confocal microscopy images showing the different mitochondrial morphology aspects from control (Ctrl), Mfn1- and Drp1-Knockdown (Kd) mouse embryonic fibroblasts. Mitochondria were labelled using an anti-TOM20 antibody. Scale bars: 10 um. Image from (PMID: 30030364). (B, C) Schematic representations of the molecular mechanisms regulating (B) mitochondrial fusion (Image from PMID: 30030364) and (C) mitochondrial division (Image from PMID: 33446409).
Cells regulate their mitochondrial network from a tubular network to a hyperfused or fragmented morphology (Figure 1A) to not only adapt to the cellular metabolic state, and in response to cellular cues but also to control cell fate decisions [1],[2]. Mitochondrial fusion allows the efficient diffusion of metabolites, proteins and sharing of mitochondrial DNA (mtDNA) and is considered as a safeguard mechanism to enhance cell survival [3]. Mitochondrial fusion is a two-step mechanism (Figure 1B), characterised first by outer mitochondrial membrane (OMM) fusion regulated by the dynamin GTPases Mitofusin-1 and -2 (Mfn1/2), and second, by inner mitochondrial membrane (IMM) fusion controlled by the dynamin-like GTPase Optic Atrophy-1 (OPA1) [4],[1]. On the other hand, mitochondrial division is involved in critical cellular functions including mitochondria quality control, cell death and immunity [5]. Mitochondrial division is a multi-step process mainly regulated by the recruitment and oligomerization of the large GTPase Dynamin-related protein 1 (Drp1) at endoplasmic reticulum (ER) contacts, by interacting with OMM-localised adaptor proteins such as mitochondrial fission factor (Mff) and mitochondrial dynamics protein of 49 and 51kDa (MiD49/51) [1],[6] (Figure 1C).
Our main research aim is to elucidate the molecular mechanisms regulating mitochondrial morphology. We employ proteomic analysis followed by biochemical and advanced microscopy methods to identify and unravel the contribution of different proteins to mitochondrial dynamics.
While the initial steps of this process are well characterised, the precise mechanisms of the final scission event are under intense investigation. We have recently described that the Trans-Golgi network (TGN)-localized small GTPase Arf1 and its effector PI(4)KIIIB generate transient pools of phosphatidylinositol 4-phosphate (PI(4)P) on Golgi-derived vesicles, which are recruited to mitochondrial constrictions and are required for the very last steps of mitochondrial division [7]. We also have highlighted a 4-way contact between mitochondria, ER, lysosomes and TGN-vesicles occurring at fission sites [7],[8]. Using confocal and super-resolution live-cell microscopy, we investigate not only how TGN-vesicles and PI4P drive mitochondrial division, but also how they are involved in other mitochondrial dynamics events. Finally, we study the relevance of these pathways to different cellular functions including cell death and survival events, cellular metabolism and in human diseases [9],[10],[11],[12],[13].
REFERENCES
- Tilokani, L., Nagashima, S., Paupe, V. & Prudent, J. (2018)
Mitochondrial dynamics: overview of molecular mechanisms
Essays Biochem 62, 341-360. doi:10.1042/EBC20170104. - Friedman, J. R. & Nunnari, J. (2014)
Mitochondrial form and function
Nature 505, 335-343. doi:10.1038/nature12985 - Wai, T. & Langer, T. (2016)
Mitochondrial Dynamics and Metabolic Regulation
Trends Endocrinol Metab 27, 105-117. doi:10.1016/j.tem.2015.12.001 - Mattie, S., Krols, M. & McBride, H. M. (2019)
The enigma of an interconnected mitochondrial reticulum: new insights into mitochondrial fusion
Curr Opin Cell Biol 59, 159-166, doi:10.1016/j.ceb.2019.05.004 - Kraus, F., Roy, K., Pucadyil, T. J. & Ryan, M. T. (2021)
Function and regulation of the divisome for mitochondrial fission
Nature 590, 57-66. doi:10.1038/s41586-021-03214-x - Prudent, J. & McBride, H. M. (2016)
Mitochondrial Dynamics: ER Actin Tightens the Drp1 Noose
Curr Biol 26, R207-209. doi:10.1016/j.cub.2016.01.009 - Nagashima, S. et al. (2020)
Golgi-derived PI(4)P-containing vesicles drive late steps of mitochondrial division
Science 367, 1366-1371. doi:10.1126/science.aax6089 - Tabara, L. C., Morris, J. L. & Prudent, J. (2021)
The Complex Dance of Organelles during Mitochondrial Division
Trends Cell Biol 31, 241-253. doi:10.1016/j.tcb.2020.12.005 - Morita, M. et al. (2017)
mTOR Controls Mitochondrial Dynamics and Cell Survival via MTFP1
Mol Cell 67, 922-935 e925. doi:10.1016/j.molcel.2017.08.013 - Janer, A. et al. (2016)
SLC25A46 is required for mitochondrial lipid homeostasis and cristae maintenance and is responsible for Leigh syndrome
EMBO Mol Med 8, 1019-1038. doi:10.15252/emmm.201506159 - Booty, L. M. et al. (2019)
Selective Disruption of Mitochondrial Thiol Redox State in Cells and In Vivo
Cell Chem Biol 26, 449-461 e448. doi:10.1016/j.chembiol.2018.12.002 - Beninca, C. et al. (2020)
Mutation in the MICOS subunit gene APOO (MIC26) associated with an X-linked recessive mitochondrial myopathy, lactic acidosis, cognitive impairment and autistic features
J Med Genet 58, 155-167. doi:10.1136/jmedgenet-2020-106861 - Pezet, M. G. et al. (2021)
Oxygen tension modulates the mitochondrial genetic bottleneck and influences the segregation of a heteroplasmic mtDNA variant in vitro
Commun Biol 4, 584, doi:10.1038/s42003-021-02069-2