SUBSTRATE BINDING AND PROTON COUPLING
In 2006, we identified the substrate binding site of mitochondrial carriers, consisting of three major contact points, which are located on the even-numbered α-helices.
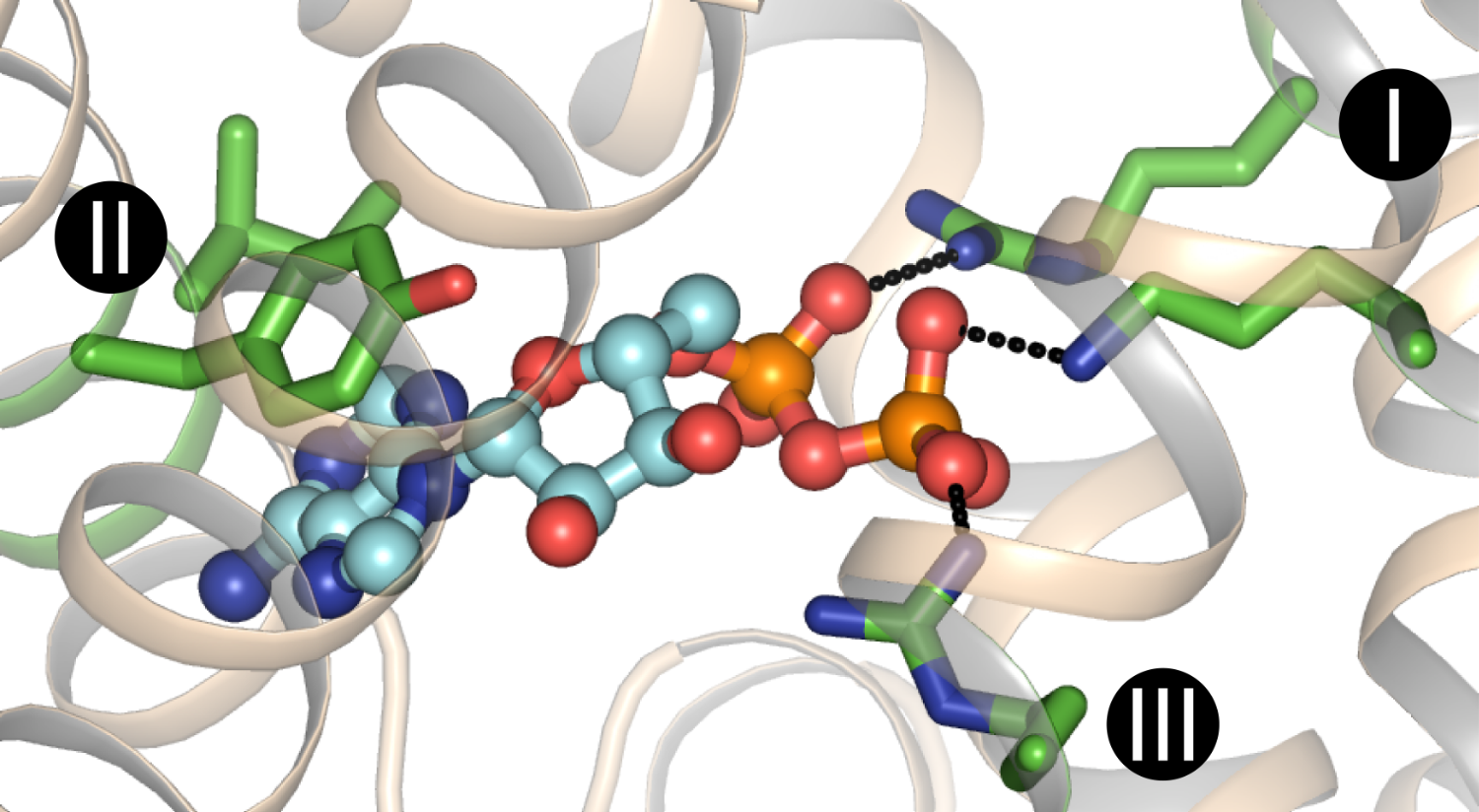
Figure The substrate binding site of the mitochondrial ADP/ATP carrrier, which was proposed to consist of glycine, isoleucine and tyrosine residues for binding of the adenine group and one lysine and two arginine residues for binding of the phosphate groups of ADP. In this way the charge is neutralised for import of ADP into mitochondria.
The substrate binding site was identified by using comparative structural models and chemical and distance constraints to identify conserved substrate binding sites that were capable of discriminating between different substrates in the amino and keto acid transporters [1][2]. The main contact points involved in substrate binding, indicated by roman numerals I, II and III, were located on the even-numbered α-helices. The second approach exploited the principle that mitochondrial carriers have a high degree of three-fold pseudo-symmetry, whereas the transported substrates are asymmetric in structure and chemistry. Therefore, the residues involved in substrate binding must deviate from each other in order to couple the binding of the asymmetric substrate to a symmetric transport mechanism [3]. In addition, conserved negatively charged asymmetric residues were observed in the substrate binding sites of carriers that transport substrates by proton coupling, indicating that proton and substrate binding occurs simultaneously [4]. Importantly, no other clusters of asymmetric residues were found on the water-accessible surfaces of the carriers, indicating that mitochondrial carriers have a single substrate binding site.
We are applying structural, biophysical and biochemical methods to study substrate binding and proton coupling in mitochondrial carriers.
REFERENCES
- Kunji ERS & Robinson AJ
The conserved substrate binding site of mitochondrial carriers.
Biochim Biophys Acta 1757, 1237-48 - Robinson AJ & Kunji ERS (2006)
Mitochondrial carriers in the cytoplasmic state have a common substrate binding site.
Proc Natl Acad Sci U S A 103, 2617-22 - Robinson AJ, Overy C & Kunji ERS(2008)
The mechanism of transport by mitochondrial carriers based on analysis of symmetry.
Proc Natl Acad Sci U S A 105, 17766-71 - Kunji ERS & Robinson AJ(2010)
Coupling of proton and substrate translocation in the transport cycle of mitochondrial carriers.
Curr Opin Struct Biol 20, 440-7