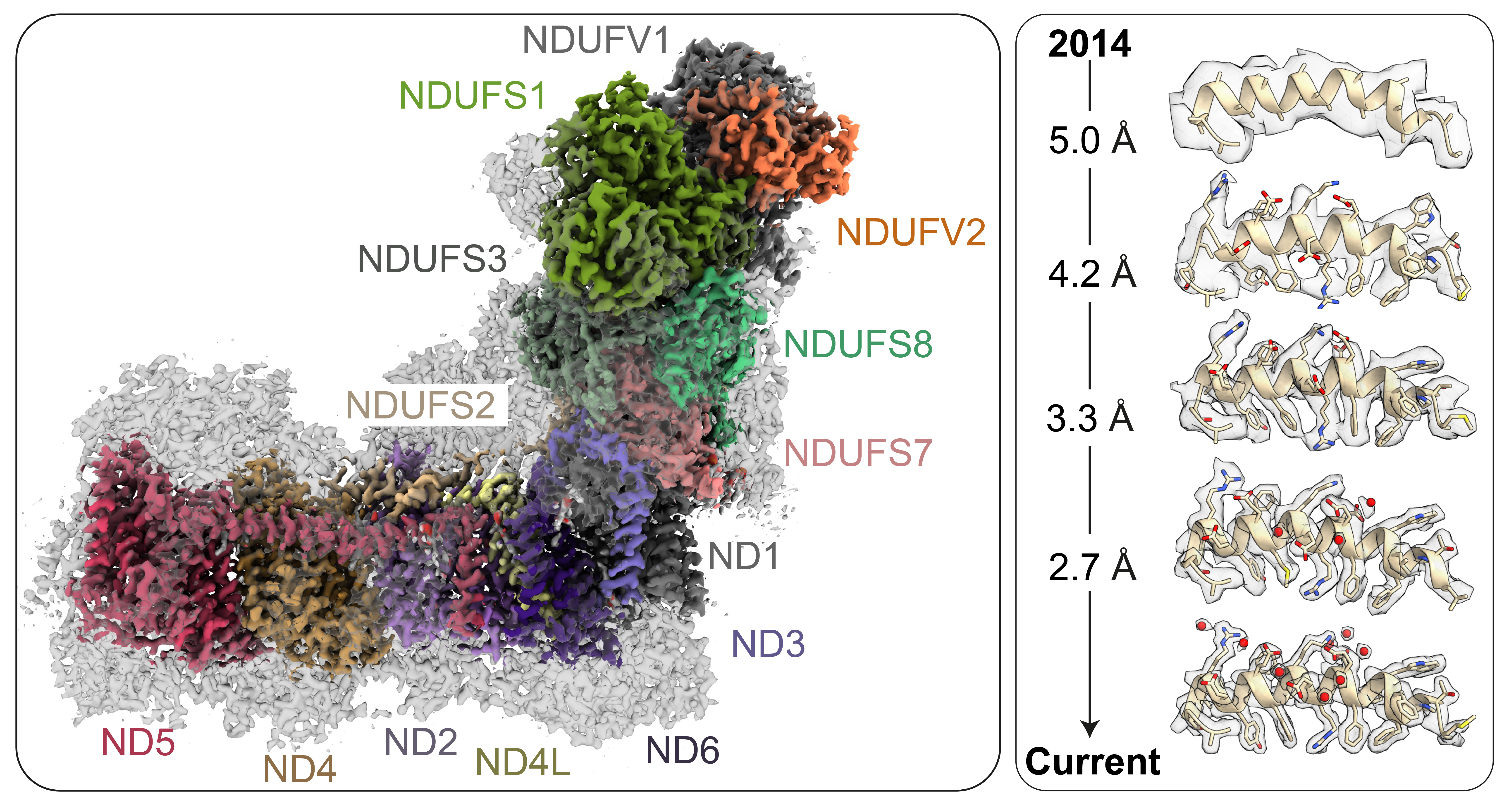
COMPLEX I cryoEM
Figure Left: Cryo-EM density map of mammalian complex I with the fourteen catalytic core subunits coloured, and the mammalian supernumerary subunits in transparent grey. Right: Progressive improvement in the resolution of our cryoEM density maps since 2014. The same helix is shown in each case, starting from a density ‘tube’ in 2014, with improving density features first attributed to individual side chains and then to water molecules.
Mammalian complex I is one of the largest enzymes in the cell. A total of ~1 MDa in mass, it contains nine electron transfer cofactors and a combination of 45 hydrophobic and hydrophilic proteins encoded on both the mitochondrial and nuclear genomes. As an irregularly shaped unstable membrane protein unsuited to crystallisation, it evaded detailed structural characterisation for many years. However, since the first mammalian cryo-EM complex I structure in 2014 [1] revealed the overall architecture of the enzyme, substantial progress has been made through both sample improvements and developments in cryoEM hardware and data-processing software. Our initial focus on improving the resolution in order to determine the first ‘complete’ structure with all the subunits and cofactors assigned and modelled [2], [3] has now shifted to defining the structures of biochemically defined [4] and inhibitor-bound states [5], [6], to learning about the details of the proton-pumping machinery by identifying water molecules and hydrogen-bonding networks [7], and to discovering the modes of action of mutations in mitochondrial disease models [8].
In the Hirst lab, cryoEM is now a central feature of all our projects that approach the function and dysfunction of complex I on a molecular level. The visualisation of structural information, providing a detailed picture of what is happening inside the protein, is crucial for allowing us to rationalise, interrogate and integrate functional, biomedical and clinical data. Using cryoEM we are able to detect and deconvolute large-scale domain movements and structural disruptions, localised catalytic and regulatory conformational changes between different states and enzyme variants, hydration and hydrogen bonding networks, and small molecule interactions. We are currently using cryo-EM as a tool to study:
- Complex I mutations in mitochondrial disease models;
- Inhibitor and drug binding to complex I;
- The mechanism of complex I catalysis;
- Biogenesis, assembly and stability of complex I.
References
- Vinothkumar KR, Zhu J & Hirst J. (2014)
Architecture of mammalian respiratory complex I.
Nature 515, 80-84 - Zhu J, Vinothkumar KR & Hirst J (2016)
Structure of mammalian respiratory complex I.
Nature 536, 354-358 - Agip A-NA, Blaza JN, Bridges HR, Viscomi C, Rawson S, Muench SP & Hirst J (2018)
CryoEM structures of complex I from mouse heart mitochondria in two biochemically-defined states.
Nat Struct Mol Biol 25, 548-556 - Blaza JN, Vinothkumar KR & Hirst J (2018)
Structure of the deactive state of mammalian respiratory complex I.
Structure 26, 312-319 - Bridges HR, Fedor JG, Blaza JN, Di Luca A, Jussupow A, Jarman OD, Wright JJ, Agip A-NA, Gamiz-Hernandez AP, Roessler MM, Kaila VRI & Hirst J (2020)
Structure of inhibitor-bound mammalian complex I.
Nat Commun 11, 5261 - Chung I, Serreli R, Cross J. B., Di Francesco M. E., Marszalek J. R. & Hirst, J (2021)
Cork-in-bottle mechanism of inhibitor binding to mammalian complex I.
Sci Adv 7, eabg4000 - Grba DN & Hirst J (2020)
Mitochondrial complex I structure reveals ordered water molecules for catalysis and proton translocation.
Nat Struct Mol Biol 27, 892-900 - Yin Z, Burger N, Kula-Alwar D, Aksentijević D, Bridges HR, Prag HA, Grba DN, Viscomi C, James AM, Mottahedin A, Krieg T, Murphy MP & Hirst J (2021)
Structural basis for a complex I mutation that blocks pathological ROS production.
Nature Commun 12, 707