PROTEIN METHYLATION
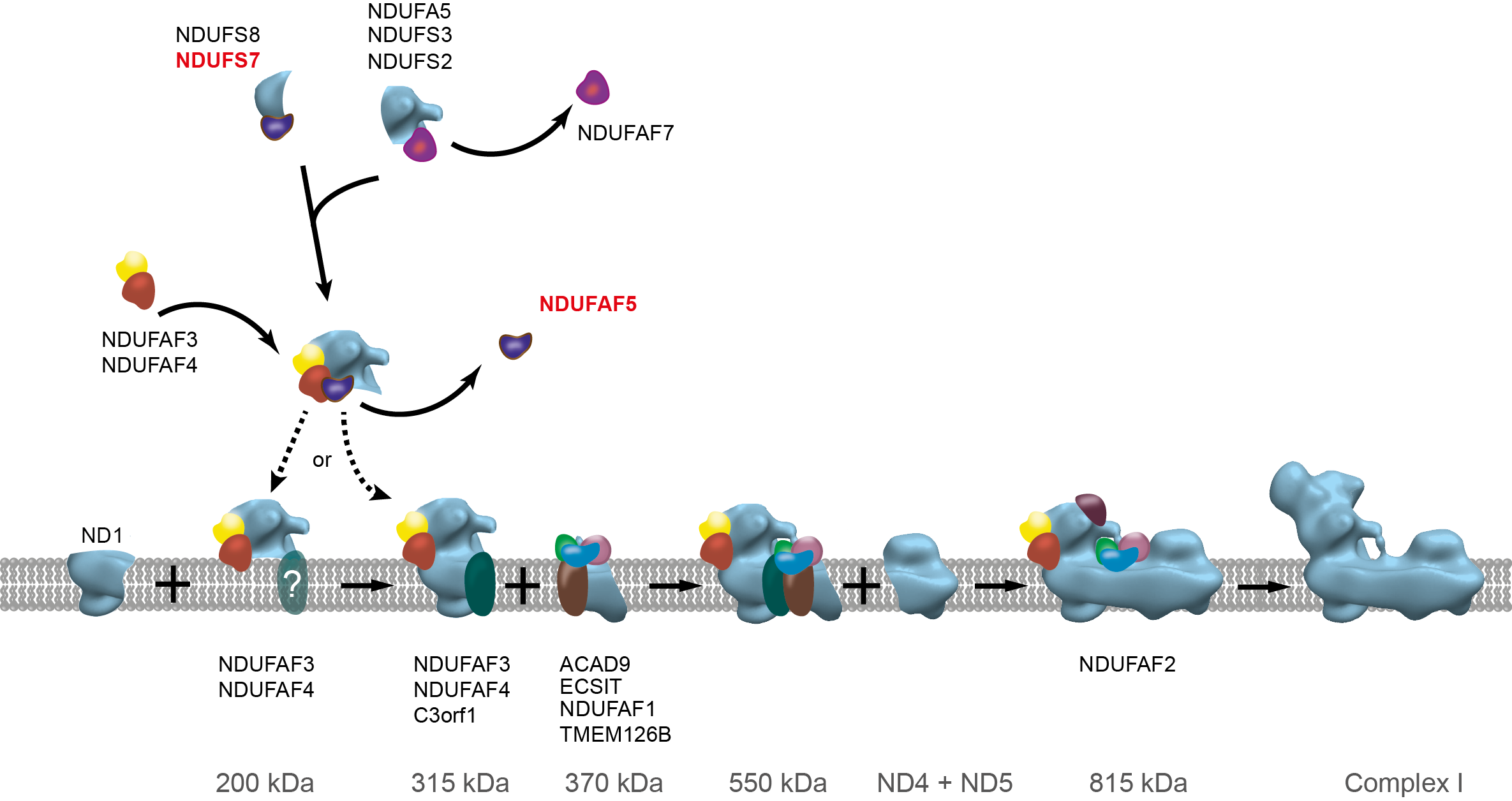
Figure Participation of NDUFAF5 in the pathway of assembly of human complex I. Assembly factors associated stably with sub-complexes in the pathway are indicated. Hydroxylation of NDUFS7 by NDUFAF5 occurs at an early stage. The scheme is derived from an earlier version, and it is not known whether the 200-kDa sub-complex is associated with the membrane or not, as indicated. The 200-kDa sub-complex joins with membrane subunit ND1 and assembly factor C3orf1, plus subunits NDUFA3, NDUFA8, and NDUFA13 to form the 315-kDa membrane-bound sub-complex.
The post-translational methylation of proteins by methyltransferases, with S-adenosylmethionine as the methyl donor, occurs primarily on the side-chains of lysine and arginine residues, but histidyl and glutamyl side chains and α-amino and α-carboxy groups can be methylated also. The ε-amino groups of lysine residues can carry one, two or three methyl groups, and the guanidino moieties of arginines can be monomethylated, and dimethylated either symmetrically or asymmetrically. It is well known that lysine residues in apocytochrome c are methylated by Ctm1p in the cytoplasm of Saccharomyces cerevisiae [PMID5459636] [1], but this modification has no clear role, and the mammalian orthologue is unmodified. Trimethyllysine residues have been characterized in three proteins isolated directly from human mitochondria; they are citrate synthase [2], ADP/ATP translocase [3] and the c-subunit in the rotor of ATP synthase [4]. A dimethylarginine residue has been characterized in the NDUFS2 subunit of complex I [5], and a complex pattern of methylation of three histidine residues has been found near to the N-terminus of the NDUFB3 subunit of complex I [6].
In our studies of methylation in human mitochondria, we have identified two methylases in the matrix of human mitochondria. NDUFAF7 methylates Arg-85 in the NDUFS2 subunit of complex I [7], and METTL20 methylates the recognition loop of the electron transfer flavoprotein [8]. Both are members of the family of 7β-strand methyltransferases. A third member of the family, NDUFAF5, also found in the mitochondrial matrix, introduces a hydroxyl group into Arg-73 in NDUFS7 at an early stage in the assembly of the complex I [9]. Recently, we and others [10] have found that FAM173B is the methyltransferase that trimethylates Lys-43 in the c-subunit of ATP synthase.
REFERENCES
- Polevoda B, Martzen MR, Das B, Phizicky EM & Sherman F (2000)
Cytochrome c methyltransferase, Ctm1p, of yeast.
J Biol Chem 275, 20508-20513 - Bloxham DP, Ericsson LH, Titani K, Walsh KA & Neurath H (1980)
Limited proteolysis of pig heart citrate synthase by subtilisin, chymotrypsin, and trypsin.
Biochemistry 19, 3979-39785 - Aquila H, Misra D, Eulitz M & Klingenberg M (1982)
Complete amino acid sequence of the ADP/ATP carrier from beef heart mitochondria.
Hoppe Seylers Z Physiol Chem 363, 345-349 - Chen R, Fearnley IM, Palmer DN & Walker JE (2004)
Lysine 43 is trimethylated in subunit C from bovine mitochondrial ATP synthase and in storage bodies associated with batten disease.
J Biol Chem 279, 21883-21887 - Carroll J, Ding S, Fearnley IM & Walker JE (2013)
Post-translational modifications near the quinone binding site of mammalian complex I.
J Biol Chem 288, 24799-24808 - Carroll J, Fearnley IM, J Skehel M, Runswick MJ, Shannon RJ, Hirst J & Walker JE (2005)
The post-translational modifications of the nuclear encoded subunits of complex I from bovine heart mitochondria.
Mol Cell Proteomics 4, 693-9 - Rhein VF, Carroll J, Ding S, Fearnley IM & Walker JE (2013)
NDUFAF7 methylates arginine 85 in the NDUFS2 subunit of human complex I.
J Biol Chem 288, 33016-26 - Rhein VF, Carroll J, He J, Ding S, Fearnley IM & Walker JE (2014)
Human METTL20 methylates lysine residues adjacent to the recognition loop of the electron transfer flavoprotein in mitochondria.
J Biol Chem 289, 24640-24651 - Rhein VF, Carroll J, Ding S, Fearnley IM & Walker JE (2016)
NDUFAF5 hydroxylates NDUFS7 at an early stage in the assembly of human complex I.
J Biol Chem 291, 14851-14860 - Małecki JM, Willemen HLDM, Pinto R, Y Y Ho A, Moen A, Kjønstad IF, Burgering BMT, Zwartkruis F, Eijkelkamp N & Falnes PØ (2019)
Lysine methylation by the mitochondrial methyltransferase FAM173B optimizes the function of mitochondrial ATP synthase.
J Biol Chem 294, 1128-1141