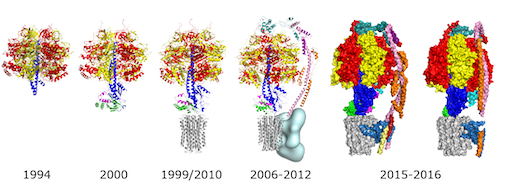
EVOLUTION OF THE STRUCTURE OF ATP SYNTHASE
| 1994 | The first atomic resolution structure of the F1 catalytic domain of the ATP synthase from bovine heart mitochondria determined by X-ray crystallography. The δ- and ε-subunits and parts of the γ-subunit were not resolved, although they were present in the crystals [1]. |
| 1999 | The structure of F1-ATPase plus the c10 ring in the membrane domain of the enzyme from Saccharomyces cerevisiae [2]. |
| 2000 | The complete structure of bovine F1-ATPase [3]. |
| 2010 | The structure of the bovine F1-c8-ring sub-complex revealed that it translocates 2.7 protons per ATP synthesised, making it the most efficient ATP synthase. The c8 ring is probably conserved throughout metazoans [4]. |
| 2006-2012 | The structure of the membrane extrinsic section of the peripheral stalk solved separately [5] and in association with F1-ATPase [6]. The presence of the peripheral stalk had been deduced earlier from reconstitution experiments with individual subunits from the central and peripheral stalks. |
| 2012 | A low resolution cryo-EM map at 17Å resolution allowed a complete mosaic structure of the bovine enzyme to be built up from high resolution structures of individual domains [7]. The high resolution part of this structure corresponded to about 85% of the entire enzyme. The remaining low resolution region in the membrane domain of the enzyme (pale blue) contains the ATP6 (or a) subunit and supernumerary subunits e, f, g, DAPIT, 6.8 proteolipid and ATP8 (or A6L). |
| 2015-2016 | Several “intermediate” resolution cryo-EM maps (at about 7 Å resolution) revealed the extraordinary and unique fold of the ATP6 subunit, with a bundle of four α-helices inclined at 30o to the plane of the membrane [8] [9] [10] [11]. This subunit and the rotating c-ring provide the transmembrane proton pathway. The positions of several supernumerary subunits were inferred from published biochemical data [12]. |
| 2015 | The structure of an intact ATP synthase from the α-proteobacterium, Paracoccus denitrificans [13], was solved at 4 Å resolution by X-ray crystallography revealing new features of the peripheral stalk and showing the same unique fold in the bacterial a-subunit as in the mitochondrial ATP6 subunit. In the structure, part of the ζ-subunit was resolved, with an α-helical structure at its N-terminus bound in a similar way to the binding of the α-helical inhibitory region of IF1 to mitochondrial enzymes. |
| 2016-present | Several structures of ATP synthases have been solved to ~3-7 Å by cryo-em. From mitochondria: the intact dimer from Yarrowia lipolytica [10], dimer and monomer from S. cerevisiae [14] [15], Polytomella sp. dimer [16], and an intact tetramer from Sus scrofa (pig) [17]; from bacteria: E. coli [18] [19]; Geobacillus stearothermophilus [20]; from plants: the chloroplast enzyme from Spinacea oleracea [21]. |
REFERENCES
- Abrahams JP, Leslie AG, Lutter R & Walker JE (1994)
Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria.
Nature 370, 621-628 - Stock D, Leslie AG & Walker JE (1999)
Molecular architecture of the rotary motor in ATP synthase.
Science 286, 1700-1705 - Gibbons C, Montgomery MG, Leslie AG & Walker JE (2000)
The structure of the central stalk in bovine F(1)-ATPase at 2.4 A resolution.
Nat Struct Biol 7, 1055-1061 - Watt IN, Montgomery MG, Runswick MJ, Leslie AGW & Walker JE (2010)
Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria.
Proc Natl Acad Sci U S A 107, 16823-16827 - Dickson VKane, Silvester JA, Fearnley IM, Leslie AGW & Walker JE (2006)
On the structure of the stator of the mitochondrial ATP synthase.
EMBO J 25, 2911-2918 - Rees DM, Leslie AGW & Walker JE (2009)
The structure of the membrane extrinsic region of bovine ATP synthase.
Proc Natl Acad Sci U S A 106, 21597-21601 - Baker LA, Watt IN, Runswick MJ, Walker JE & Rubinstein JL(2012)
Arrangement of subunits in intact mammalian mitochondrial ATP synthase determined by cryo-EM.
Proc Natl Acad Sci U S A 109, 11675-11680 - Allegretti M, Klusch N, Mills DJ, Vonck J, Kühlbrandt W & Davies KM (2015)
Horizontal membrane-intrinsic α-helices in the stator a-subunit of an F-type ATP synthase.
Nature 521, 237-240 - Zhou A, Rohou A, Schep DG, Bason JV, Montgomery MG, Walker JE, Grigorieff N & Rubinstein JL (2015)
Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife 4, e10180 - Hahn A, Parey K, Bublitz M, Mills DJ, Zickermann V, Vonck J, Kühlbrandt W & Meier T (2016)
Structure of a Complete ATP Synthase Dimer Reveals the Molecular Basis of Inner Mitochondrial Membrane Morphology.
Mol Cell 63, 445-456 - Vinothkumar KR, Montgomery MG, Liu S & Walker JE (2016)
Structure of the mitochondrial ATP synthase from Pichia angusta determined by electron cryo-microscopy.
Proc Natl Acad Sci U S A 113, 12709-12714 - Lee J, Ding S, Walpole TB, Holding AN, Montgomery MG, Fearnley IM & Walker JE (2015)
Organization of Subunits in the Membrane Domain of the Bovine F-ATPase Revealed by Covalent Cross-linking.
J Biol Chem 290, 13308-13320 - Morales-Rios E, Montgomery MG, Leslie AGW & Walker JE (2015)
Structure of ATP synthase from Paracoccus denitrificans determined by X-ray crystallography at 4.0 Å resolution.
Proc Natl Acad Sci U S A 112, 13231-13236 - Guo H, Bueler SA & Rubinstein JL (2017)
Atomic model for the dimeric F region of mitochondrial ATP synthase.
Science 358, 936-940 - Srivastava AP, Luo M, Zhou W, Symersky J, Bai D, Chambers MG, Faraldo-Gómez JD, Liao M & Mueller DM (2018)
High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane.
Science 360, eaas9699 - Murphy BJ, Klusch N, Langer J, Mills DJ, Yildiz Ö & Kühlbrandt W (2019)
Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F-F coupling.
Science 364, eaaw9128 - Gu J, Zhang L, Zong S, Guo R, Liu T, Yi J, Wang P, Zhuo W & Yang M (2019)
Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science 364, 1068-1075 - Sobti M, Smits C, Wong ASw, Ishmukhametov R, Stock D, Sandin S & Stewart AG (2016)
Cryo-EM structures of the autoinhibited E. coli ATP synthase in three rotational states.
Elife 5, e21598 - Sobti M, Ishmukhametov R, Bouwer JC, Ayer A, Suarna C, Smith NJ, Christie M, Stocker R, Duncan TM & Stewart AG (2019)
Cryo-EM reveals distinct conformations of ATP synthase on exposure to ATP.
Elife 8, e43864 - Guo H, Suzuki T & Rubinstein JL (2019)
Structure of a bacterial ATP synthase.
Elife 8, 43128 - Hahn A, Vonck J, Mills DJ, Meier T & Kühlbrandt W (2018)
Structure, mechanism, and regulation of the chloroplast ATP synthase.
Science 360, eaat4318