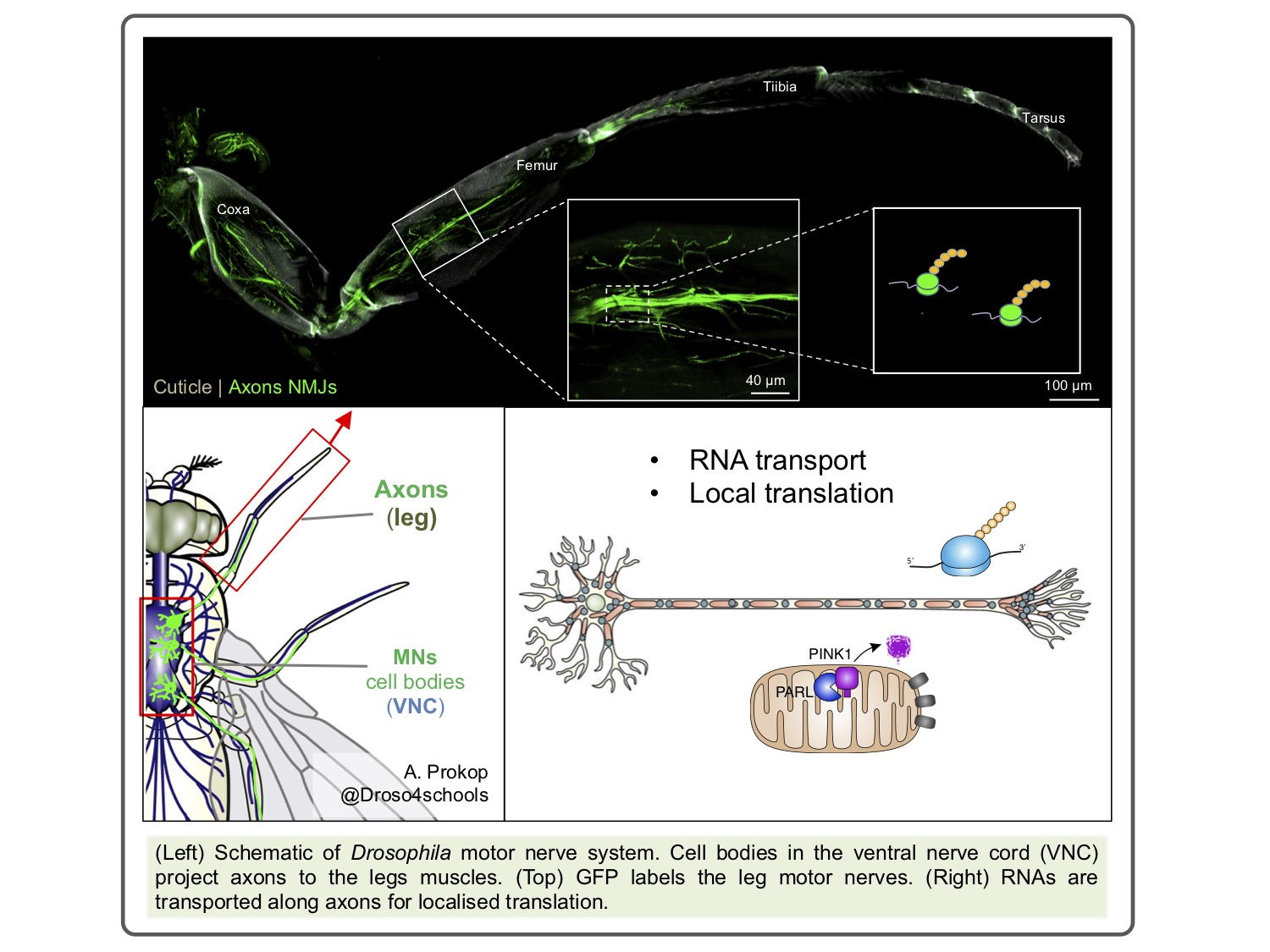
REGULATION OF RNA IN MITOCHONDRIAL AND NEURONAL HOMEOSTASIS
The elaborate architecture of neurons presents extraordinary challenges for molecular cell biology processes, where sites of intense activity (e.g. synapses) may be great distances from the cell bodies. The high energy demands of synapses put an extra burden on mitochondrial maintenance. The extraordinary length of some neurons renders the local supply of component proteins a practical necessity. This is achieved by the regulated axonal transport of mRNAs and localised translation[1].
Several neurodegenerative diseases selectively affect long nerves, for example amyotrophic lateral sclerosis (ALS), and are associated with defective axonal transport[2] Functional studies of mutations that cause the disease have begun to implicate a number of mechanisms. One key mechanism that is emerging is the dysregulation of RNA biology[3],[4]. Current evidence indicates that multiple aspects of mRNA formation and handling are affected in disease conditions, including expression, maturation, stability, subcellular distribution and translation. It is unclear why mRNA handling leads specifically to the demise of motor nerves to cause ALS, but the observed ‘dying-back’ pathology also supports that cellular disruption begins at the distal nerve terminus[5]. We hypothesise that disruptions to the myriad of normal RNA regulatory processes leads to altered populations of mRNAs available for translation in motor nerve termini in ALS. However, significant technical challenges have meant that this phenomenon is poorly understood in an in vivo setting.
To explore this idea, we are investigating which mRNA molecules are actively undergoing translation (translatome) specifically in the axons and synapses of motor nerves in vivo. We have established a method to extract tagged ribosomes specifically from motor nerve axons[6]. Thus, we seek to understand the MN axonal translatome under normal conditions over the fly life-course, investigate how axonal and somal translation is affected during disease progression, and determine the relative contribution of different dysregulated axonal mRNAs to pathogenicity by assessing their functional impact on the disease-relevant circuits at an organismal level. Results delineated from this project will provide new insights into how dysregulated RNA biology specifically impacts on motor neuron survival.
REFERENCES
- Holt CE & Schuman EM (2013)
The central dogma decentralized: new perspectives on RNA function and local translation in neurons.
Neuron 80, 648-657 - Baldwin KR, Godena VK, Hewitt VL & Whitworth AJ (2016)
Axonal transport defects are a common phenotype in Drosophila models of ALS.
Hum Mol Genet 25, 2378-2392 - Peters OM, Ghasemi M & Brown RH (2015)
Emerging mechanisms of molecular pathology in ALS.
J Clin Invest 125, 1767-1779 - J. Taylor P, Brown RH & Cleveland DW (2016)
Decoding ALS: from genes to mechanism.
Nature 539, 197-206 - Dadon-Nachum M, Melamed E & Offen D (2011)
The "dying-back" phenomenon of motor neurons in ALS.
J Mol Neurosci 43, 470-477 - Shigeoka T, Jung J, Holt CE & Jung H (2018)
Axon-TRAP-RiboTag: Affinity Purification of Translated mRNAs from Neuronal Axons in Mouse In Vivo.
Methods Mol Biol 1649, 85-94