With respect to Douglas Adams:
Metabolism is complicated. Really complicated. You just won't believe how vastly, hugely, mind-bogglingly complex it is. I mean, you may think the citric acid cycle is complicated, but that's just peanuts to what's happening in the cell.
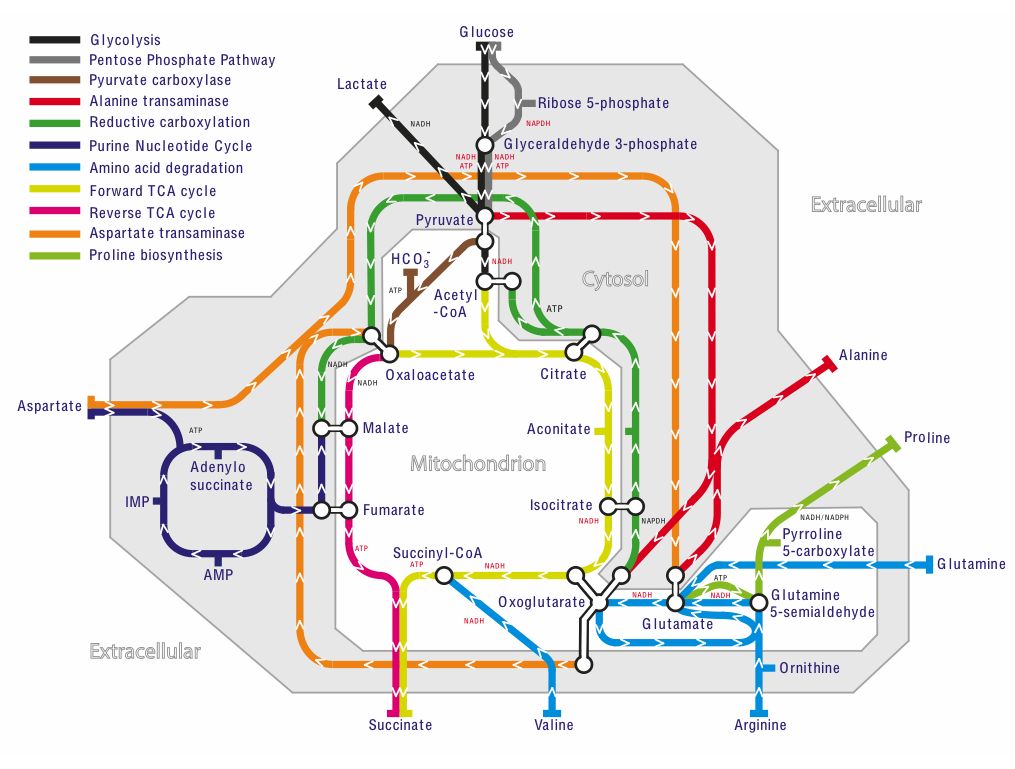
Mitochondrial metabolism is complicated and arises from dynamic networks of thousands of genes, proteins, and metabolites. Challenges to understanding the mechanisms of mitochondrial diseases are their huge variability in clinical symptoms and penetrance, and their wide genetic and biochemical differences.
Because the network is so complicated, we build computers models of these networks [1],[2]. We then use these models to simulate mitochondrial metabolism under normal conditions, and also what happens to energy production and metabolism if we change the activity of an enzyme, or the availability of a metabolite. These changes model what happens when mitochondrial proteins are affected by genetic mutations as in mitochondrial diseases [3],[4], or the supply of metabolites is increased or decreased, as in heart disease [5], ischaemia/reperfusion injury [6], diabetes [7], or cancer. If a change causes a big drop in energy production, then it is likely to cause serious disease . But sometimes the metabolic network can re-route around a disease gene to limit the damage. If we can bolster that pathway, then it may be a therapy that reduces the burden of the disease.
Our models are the most comprehensive of their type available - with over 400 reactions - enabling a systems-level view of metabolism and interpreting biochemical and clinical data. Our latest model, MitoCore, is available here.
REFERENCES
-
Smith, AC & Robinson, AJ (2011)
A metabolic model of the mitochondrion and its use in modelling diseases of the tricarboxylic acid cycle.
BMC Syst Biol 5, 102 - Zieliński, ŁP, Smith, AC, Smith, AG & Robinson, AJ (2016)
Metabolic flexibility of mitochondrial respiratory chain disorders predicted by computer modelling.
Mitochondrion 31, 45-55 - Majd, H, King, MS, Smith, AC & Kunji, ERS (2018)
Pathogenic mutations of the human mitochondrial citrate carrier SLC25A1 lead to impaired citrate export required for lipid, dolichol, ubiquinone and sterol synthesis.
Biochim Biophys Acta 1859, 1-7 - Ashrafian, H, Czibik, G, Bellahcene, M, Aksentijević, D, Smith, AC, Mitchell, SJ, Dodd, MS, Kirwan, J, Byrne, JJ, Ludwig, C, Isackson, H, Yavari, A, Støttrup, NB, Contractor, H, Cahill, TJ, Sahgal, N, Ball, DR, Birkler, RID, Hargreaves, I, Tennant, DA, Land, J, Lygate, CA, Johannsen, M, Kharbanda, RK, Neubauer, S, Redwood, C, de Cabo, R, Ahmet, I, Talan, M, Günther, UL, Robinson, AJ, Viant, MR, Pollard, PJ, Tyler, DJ & Watkins, H (2012)
Fumarate is cardioprotective via activation of the Nrf2 antioxidant pathway.
Cell Metab 15, 361-371 - Chouchani, ET, Pell, VR, Gaude, E, Aksentijević, D, Sundier, SY, Robb, EL, Logan, A, Nadtochiy, SM, Ord, ENJ, Smith, AC, Eyassu, F, Shirley, R, Hu, C-H, Dare, AJ, James, AM, Rogatti, S, Hartley, RC, Eaton, S, Costa, ASH, Brookes, PS, Davidson, SM, Duchen, MR, Saeb-Parsy, K, Shattock, MJ, Robinson, AJ, Work, LM, Frezza, C, Krieg, T & Murphy, MP (2014) Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 515, 431-435
- Yang, L, Vaitheesvaran, B, Hartil, K, Robinson, AJ, Hoopmann, MR, Eng, JK, Kurland, IJ & Bruce, JE (2011)
The fasted/fed mouse metabolic acetylome: N6-acetylation differences suggest acetylation coordinates organ-specific fuel switching.
J Proteome Res 10, 4134-4139 - Smith, AC, Eyassu, F, Mazat, J-P & Robinson, AJ (2017)
MitoCore: a curated constraint-based model for simulating human central metabolism.
BMC Syst Biol 11, 114