Submitted by Penny Peck on Mon, 05/12/2022 - 11:45
The development of curative treatments for mitochondrial diseases, which are often caused by mutations in mitochondrial DNA (mtDNA) that impair energy metabolism and other aspects of cellular homoeostasis, is hindered by an incomplete understanding of the underlying biology and a scarcity of cellular and animal models.
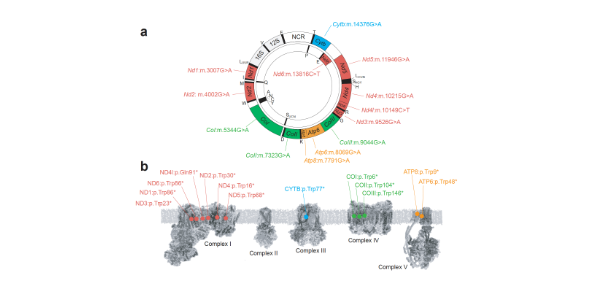
Dr Michal Minczuk and his colleagues in our Mitochondrial Genetics research group report in Nature Biomedical Engineering the design and application of a library of double-stranded-DNA deaminase-derived cytosine base editors (DdCBEs) optimised for the precise ablation of every mtDNA protein-coding gene in the mouse mitochondrial genome.
The library, which these researchers have named MitoKO, can be used to produce near-homoplasmic knockout cells in vitro and to generate a mouse knockout with high heteroplasmy levels and no off-target edits.
MitoKO should facilitate systematic and comprehensive investigations of mtDNA-related pathways and their impact on organismal homoeostasis, and aid the generation of clinically meaningful in vivo models of mtDNA dysfunction.
Full publication reference:
Pedro Silva-Pinheiro, Christian D. Mutti, Lindsey Van Haute, Christopher A. Powell, Pavel A. Nash, Keira Turner & Michal Minczuk. (2022) A library of base editors for the precise ablation of all protein-coding genes in the mouse mitochondrial genome. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-022-00968-1

